Antibiotics resistance of harmful organisms becomes a major problem – in agriculture, in human and veterinary medicine. Scientists now try to come up with new methods. They think that CRISPR (discovered less than 10 years ago) might be a solution.
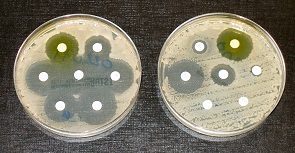
Scientists don’t often highlight it, but resistance of harmful organisms to our drugs is becoming a major threat to modern lifestyle. Plants have developed resistance to even the most powerful weed killers, like in the amaranth that threatens US cotton farms. As for human healthcare, microorganisms are very versatile and quickly develop defences against the bombardments with antibiotics with which we treat them. Many antibiotic cures prescribed are even pointless. Maybe not surprising, because it may be hard for a doctor to refuse the patient a treatment that might cure him (or her). But the overall effect is the quick development of antibiotics resistance.
Antibiotics resistance develops fast
The New York Times recently highlighted that an increasing number of bacteria now has become resistant to one or more antibiotics. Each year roughly 700,000 people around the world die from infections by resistant bacteria; and by 2050 the number could rise to 10 million, according to United Nations estimates. And viruses have a habit of making themselves invisible to medicines by hiding inside host cells. A major cause of antibiotics resistance is the coarseness of most therapies. Broad-spectrum antibiotics will kill most bacteria they find on their path, including the beneficial ones. This is a good incentive for bacteria to develop resistant strains. And in the past two decades, scientists have not discovered any new families of antibiotics. Are we running out of resources?
Scientists are now turning to CRISPR for new defence mechanisms. Originally, CRISPR together with its related Cas enzymes forms a bacterial defence mechanism against viral infections. The bacterium stores small chunks of the viral DNA in its own genome (the CRISPR). If it is attacked again by this virus, it sends molecular scissors on them (the Cas) that disarm the attacker. Hence, CRIPR-Cas is a highly selective mechanism, quite unlike antibiotics or – in another realm – weed killers like Roundup. Therefore a scientist like David Edgell looks upon CRISPR as the next step in antimicrobial therapy. He is a biologist at the Western University in London, Ontario, and the lead author of a study published recently in Nature Communications. The scientists succeeded in controlling toxic Salmonella bacteria by a Cas enzyme carried by ordinary E.coli bacteria. Yeah, but so far this has only been done in a petri dish. It may take ten or twenty years before CRISPR-based pills will actually be administered to Salmonella infected patients.
Spare beneficial microorganisms
The major advantage of CRISPR-based cures would be that they just attack specific pathogens. And therefore spare beneficial microorganisms. Eradicating the good together with the bad bacteria may cause problems, for instance for people with weakened immune systems.
In another line of research, scientists try to use the CRISPR mechanism to target genes in microorganisms that lend antibiotics resistance to them. One team of scientists has already succeeded in isolating (and destroying) antibiotics-resistant bacteria that cause pneumonia and malaria. Others have successfully removed an antibiotics resistance gene from the Enterococcus faecalis bacterium. E. faecalis is part of the normal, benign intestinal flora, but when antibiotics kill off beneficial intestinal flora, E. faecalis can become pathogenic. Antibiotic resistant E. faecalis infections have become a major problem in hospitals. This mechanism of removing the genes responsible for resistance may extend the lifetime of our antibiotics: through the CRISPR mechanism we might specifically target resistant bacterial strains.
A daunting task still to be done
And the CRISPR mechanism could also be used against pathogenic viruses. Much like the original use of this mechanism by bacteria. Scientists have developed a CRISPR-Cas mechanism against the influenza A virus, the lymphocytic choriomeningitis virus and the vesicular stomatitis virus. Fairly mild viruses so far, but there is hope that this could also be effective against more deadly viral infections. Compared to current antiviral drugs, CRISPR has the advantage of being easy to modify as needed. If a virus evolves and mutates, the researchers can simply change the system to match whatever the virus is doing.
There is still a daunting task to be done: show that CRISPR therapies are not just viable in the lab, but also work on humans. And that we will be able to afford them. ‘We’re not ready for clinical prime time yet,’ said Dr. Rodolphe Barrangou, a microbiologist at North Carolina State University, to the New York Times. ‘But we’re getting there.’
Interesting? Then also read:
Welcome to the age of CRISPR
Antibiotics resistance, do ants have the solution?
Can we engineer life? Precision modification of the genome
